More Information
Submitted: April 15, 2021 | Approved: April 23, 2021 | Published: April 26, 2021
How to cite this article: Luisetto M, Tarro G, Khan FA, Edbey K, Mashori GR, et al. RBD targeted COVID vaccine and full length spike-protein vaccine (mutation and glycosylation role) relationship with procoagulant effect. J Child Adult Vaccines Immunol. 2021; 5: 001-008.
DOI: 10.29328/journal.jcavi.1001007
Copyright License: © 2021 Luisetto M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: RBD domain; Spike full length; Mutation; Glycosilation; COVID-19 vaccine; ADR; Thrombosys
RBD targeted COVID vaccine and full length spike-protein vaccine (mutation and glycosylation role) relationship with procoagulant effect
Luisetto M1*, Tarro G2, Farhan Ahmad Khan3, Khaled Edbey4, Mashori GR5, Yesvi AR6 and Latyschev OY7
1IMA Academy Research Centre, Applied Pharmacologist, Natural Science, Branch, 29121, Italy
2President of the T & L de Beaumont Bonelli Foundation for Cancer Research, Naples, Italy
3Professor Department of Pharmacology J.N. Medical College, AMU, Aligarh
4Professor, Department of Chemistry, University of Benghazi, Libya
5Profesor, Department of Medical & Health Sciences for Woman, Peoples University of Medical and Health Sciences for Women, Pakistan
6Founder and President, Yugen Research Organization; Undergraduate Student, Western Michigan University, MI, USA 49008
7President, IMA Academy, RU
*Address for Correspondence: Luisetto M, IMA Academy Research Centre, Applied Pharmacologist, Natural Science, Branch, 29121, Italy, Email: [email protected]
Related COVID vaccine production many different strategies was followed by the producers.
Observing some rare event of thrombosis after some COVID-19 vaccination, it is interesting to verify if the Target used for the manufacturing can be involved in a different procoagulant activity or not.
Some vaccine are suspended in some country or under a deep new verify- investigation by the regulatory agency. (EU or USA).
This fact it is relevant.
The target SPIKE-PROTEIN FULL LENGTH modified or not or towards the RBD domain can be a relevant factor.
According the article published by Oxford University 15 April 2021: Risk of rare blood clotting higher for COVID-19 than for vaccines:
“Researchers at the University of Oxford have today reported that the risk of the rare blood- clotting known as cerebral venou-s thrombosis (CVT) following COVID-19 infection is around 100 times greater than normal, several times higher than it is post-vaccination or following influenza.
The study authors, led by Professor P Harrison and Dr. M Taquet from Oxford University’s Department of Psychiatry and the NIHR Oxford Health Biomedical Research Centre, counted the number of CVT cases diagnosed in the two weeks following diagnosis of COVID-19, or after the first dose of a vaccine. The then compared these to calculated incidences of CVT following influenza, and the background level in the general population.
They report that CVT is more common after COVID-19 than in any of the comparison groups, with 30% of these cases occurring in the under 30s. Compared to the current COVID-19 vaccines, this risk is between 8-10 times higher, and compared to the baseline, approximately 100 times higher.
The breakdown comparison for reported cases of CVT in COVID-19 patients in comparison to CVT cases in those who received a COVID-19 vaccine is:
In this study of over 500,000 COVID-19 patients, CVT occurred in 39 in a million patients.
In over 480,000 people receiving a COVID-19 mRNA vaccine (Pfizer or Moderna), CVT occurred in 4 in a million.
CVT has been reported to occur in about 5 in a million people after first dose of the AZ-Oxford COVID-19 vaccine.
Compared to the mRNA vaccines, the risk of a CVT from COVID-19 is about 10 times greater.
Compared to the AZ-Oxford vaccine, the risk of a CVT from COVID-19 is about 8 times greater.”
(https://www.ox.ac.uk/news/2021-04-15-risk-rare-blood-clotting-higher-COVID-19-vaccines)
And according Tobaiqy, M.; Elkout, H.;MacLure, K. Analysis of Thrombotic Adverse Reactions of COVID-19
AstraZeneca Vaccine Reported to EudraVigilance Database. Vaccines 2021, 9, 393. https://doi.org/10.3390/vaccines9040393
“The development of safe, effective, affordable vaccines against COVID-19 remains the cornerstone to mitigating this pandemic. Early in December 2020, multiple research groups had designed potential vaccines. From 11 March 2021, several European countries temporarily suspended the use of the Oxford–AstraZeneca vaccine amid reports of blood clot events and the death of a vaccinated person, despite the European Medicines- Agency (EMA) and the World Health Organization’s assurance that there was no indication that vaccination was linked. This study aimed to identify and analyse the thrombotic adverse- reactions associated with the Oxford–AstraZeneca vaccine. This was a retrospective descriptive study using spontaneous reports submitted to the Eudra-Vigilance database in the period from 17 February to 12 March 2021. There were 54,571 adverse reaction reports, of which 28 were associated with thrombotic adverse reactions. Three fatalities were related to pulmonary embolism; one fatality to thrombosis.
With 17 million people having had the AstraZeneca vaccine, these are extremely rare events The EMA’s Pharmacovigilance Risk Assessment Committee (18 March 2021) concluded that the vaccine was safe, effective and the benefits outweighed the risks.
Conducting further analyses based on more detailed thrombotic adverse event reports, including patients’ characteristics and comorbidities, may enable assessment of the causality with higher specificity” and in article: Thrombosis after COVID-19 vaccination BMJ 2021; 373 doi: https://doi.org/10.1136/bmj.n958 (Published 14 April 2021) Cite this as: BMJ 2021;373:n958 was reported:
“Regulatory bodies have also issued warnings to the patients and healthcare professionals to be vigilant and seek prompt medical assistance if they experienced typical symptoms of cerebral venous sinus thrombosis (CVST), a potentially fatal clot in the brain [2,3]. European Medicines Agency has also revised the summary of product characteristics and listed thrombocytopenia (very low platelets) as a ‘common’ side effect (i.e., 1 in 100 to 1 in 10) of Vaxzevria, i.e., the COVID vaccine AstraZeneca [4]. The pharmacovigilance data also suggests that thrombocytopenia is also a frequent observation followed by mRNA COVID vaccines such as Pfizer or Moderna.”
Even if this are rare event, (vaccine-induced thrombosis, was “only” a few dozen out of millions of vaccinated people;The incidence of thrombosis lies between 0.2 and 0.5 per 100 000) some COVID vaccine was in this days deeply investigated by regulatory agency to verify if present a relationship or not (between thrombosis and vaccine) and in some countries two limitation of use was recently adopted.
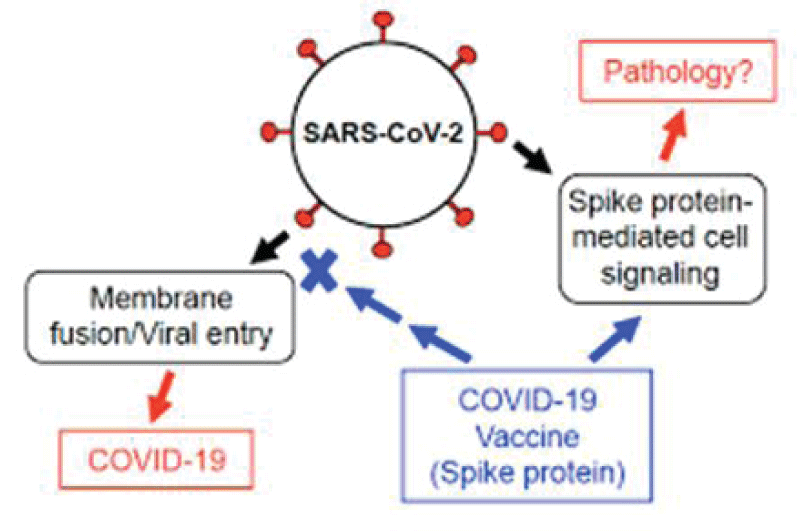
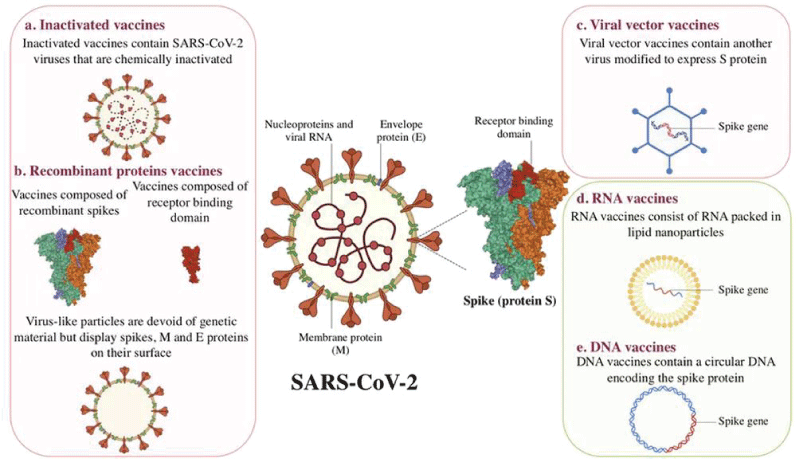
For the scope os this work It is interesting to observe the role played by spike COVID-19 protein in the cells viral entry phases but also related the vaccine production: (Figure 1).
Figure 1: SARS-CoV-2 Spike Protein Elicits Cell Signaling in Human Host Cells: Implications for Possible Consequences of COVID-19 Vaccines by Yuichiro J. Suzuki 1,*OrcID andSergiy G. Gychka if we observe the various production strategy that involve the SPIKE COVID-19 protein it is interesting to verify that:
As reported by By Lakshmi Supriya, PhD: https://www.news-medical.net/news/20210409/Oxford-AstraZeneca-COVID-vaccine-induces-cell-spikes-similar-to-SARS-CoV-2s.aspx Oxford-AstraZeneca COVID vaccine induces cell spikes similar to SARS-CoV-2’s “The virus spike-protein is the main target of vaccines, but vaccines use different methods to target the spike-protein. The Moderna and the Pfizer vaccines encode the full-length spike-protein with two mutations for stability. Sinovac’s vaccine uses an inactivated virus that presents the wild-type spike-protein.
Most vaccines aim to elicit a robust immune response, mainly against the receptor-binding-domain (RBD) of the spike-protein that has several neutralizing epitopes. To enable this, many vaccines include mutations that ensure the spike-protein is in the conformation before it fuses with the host- cell.
The AstraZeneca ChAdOx1 vaccine uses a chimpanzee- adenovirus and encodes the full-length spike-protein. It has been shown to elicit a robust immune response as well as a T-cell response. In a recent study, researchers report the characteristics of the spike-protein expressed by the vaccine.”
The virus spike-protein also has glycans, a type of sugar, coating it, which disguise the virus so that the host immune response does not detect the virus. This is a common strategy used by many viruses to elude the host immune- system. So it is crucial that the spike-proteins produced by vaccination also produce these glycans to ensure complete mimicking of the virus for the production of suitable neutralizing- antibodies.
Test for the presence of glycans, the team used HEK293 cells infected with the vaccine and used enzymes to generate glycopeptides. Tests revealed a high level of glycans present on the spike, providing evidence that the spike-proteins produced by ChAdOx1 vaccination were similar to the spike-protein expressed by natural infection, triggering an immune -response that can protect against COVID-19.
Thus, the results reveal that vaccination by ChAdOx1 produces spike-protein that is very similar to that produced by the SARS-CoV-2 virus upon natural infection, providing more evidence that the vaccine is triggering the immune- system to fight COVID-19.”
Many literature reported fatal thrombotic event in many COVID cases and the same the use of heparin is suggest to prevent this phenomena.
This increased procoagulant activity ca be related or to the body response towards the virus or due by the virus itself or by a its parts.
According Si Zhang, et al: “SARS-CoV-2 and its Spike-protein directly enhanced platelet activation such as platelet aggregation, PAC-1 binding, CD62P expression, α granule secretion, dense granule release, platelet spreading, and clot retraction in vitro, and thereby Spike-protein enhanced thrombosis formation in wild-type mice transfused with hACE2 transgenic platelets, but this was not observed in animals transfused with wild-type platelets in vivo. Further, we provided evidence suggesting that the MAPK pathway, downstream of ACE2, mediates the potentiating role of SARS-CoV-2 on platelet activation, and that platelet ACE2 expression decreases following SARS-COV-2 stimulation. SARS-CoV-2 and its Spike-protein directly stimulated platelets to facilitate the release of coagulation -factors, the secretion of inflammatory factors, and the formation of leukocyte–platelet aggregates”.
Lee Makowski, et al: “Although ACE2 (angiotensin converting enzyme 2) is considered the primary receptor for CoV-2 cell entry, recent reports suggest that alternative pathways may contribute. This paper considers the hypothesis that viral binding to cell-surface integrins may contribute to the high infectivity and widespread extra-pulmonary impacts of the SARS-CoV-2 virus. This potential is suggested on the basis of the emergence of an RGD (arginine-glycine-aspartate) sequence in the receptor-binding-domain of the spike-protein. RGD is a motif commonly used by viruses to bind cell-surface integrins. Numerous signaling- pathways are mediated by integrins and virion binding could lead to dysregulation of these pathways, with consequent tissue damage. Integrins on the surfaces of pneumocytes, endothelia-l cells and platelets may be vulnerable to CoV-2 virion binding. For instance, binding of intact virions to integrins on alveolar -cells could enhance viral entry. Binding of virions to integrins on endothelial cells could activate angiogenic cell signaling- pathways; dysregulate integrin-mediated signaling pathways controlling developmental processes; and precipitate endothelial activation to initiate blood- clotting.
Such a procoagulant state, perhaps together with enhancement of platelet aggregation through virions binding to integrins on platelets, could amplify the production of microthrombi that pose the threat of pulmonary thrombosis and embolism, strokes and other thrombotic consequences. The susceptibility of different tissues to virion–integrin interactions may be modulated by a host of factors, including the conformation of relevant integrins and the impact of the tissue micro-environment on spike-protein conformation. Patient-specific differences in these factors may contribute to the high variability of clinical presentation. There is danger that the emergence of receptor-binding-domain mutations that increase infectivity may also enhance access of the RGD motif for integrin- binding, resulting in viral strains with ACE2 independent routes of cell entry and novel integrin-mediated biological and clinical- impacts. The highly infectious variant, B.1.1.7 (or VUI 202012/01), includes a receptor-binding-domain amino acid replacement, N501Y, that could potentially provide the RGD motif with enhanced access to cell-surface integrins, with consequent clinical impacts” (Figure 2).
Figure 2: Structure of known integrin-binding proteins: (A) Virion proteins known to bind integrins through an RGD motif (shown in space-fill) include (right) foot and mouth disease virus capsid protein (5 neu—this RGD motif is highly flexible prior to integrin-binding, but structurally stabilized when bound to integrin—image is from a co-crystal of the capsid -protein and integrin with integrin structure removed to make visible the RGD -domain); (left) African horse sickness virus (1 ahs—top domain of capsid protein VP7). (B) Other proteins known to bind integrin through an RGD motif: thrombospondin (1 u x 6); prothrombin (3u69); rhodostomin (4 rqg) and triflavin (1 j 2 l) are disintegrins, small toxins from snake venom with high affinity to integrins; and fibronectin (1 fnf—domains 6–10)—an extracellular matrix protein with an integrin-binding RGD motif in its 10th domain.
Whit an observational approach some relevant literature is reported and analized to submit a final global conclusion related to the aim of this study.
All literature come from PUB MED database or other OPEN LITERATURE.
From literature
FROM nature reviews immunology https://www.nature.com/articles/s41577-020-00480-0/tables/1
Table 1 Current vaccines under development and their major antigen targets.
From: Viral targets for vaccines against COVID-Gamaleya Research Institute VACCINE, Russia Virus vector (Ad26 and Ad5) Full-length S.
Induction of RBD-specific IgG and nAbs in humans, with nAb titres similar to convalescent plasma; induction of IFNγ-associated T cell responses.
2)Virus vector (ChAdOx1)Full-length S University of Oxford, with AstraZeneca, UK Induction of S-specific IgG and nAbs in mice and NHPs; induction of high TH1 cell responses but low TH2 cell responses in mice; induction of S-specific IgG and nAbs in humans, with nAb titres similar to convalescent plasma [1] (Figure 3).
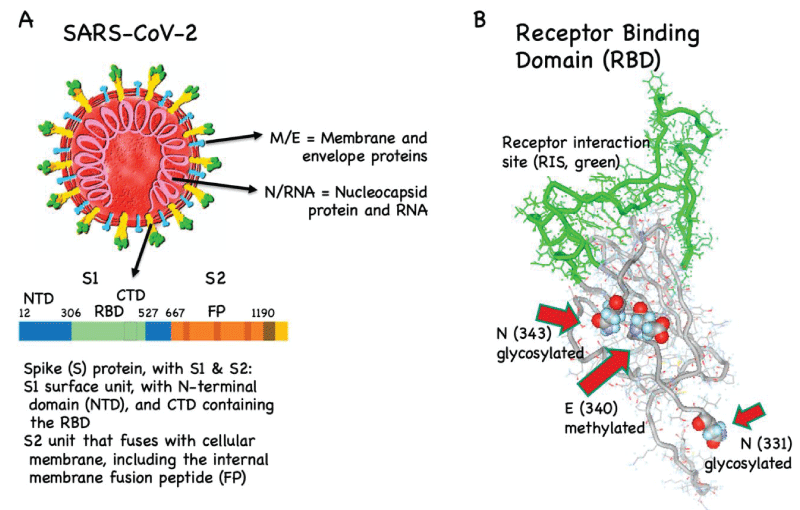
Figure 3: SARS-CoV-2, the spike (S) protein and its receptor binding-domain (RBD). (A) Coronaviruses have their name because they are decorated by prominent S proteins (yellow/green). It is the only viral protein that interacts with host cells and is the most diverging protein between different coronaviruses, particularly in its receptor binding-domain (RBD, green). RBD binds to angiotensin converting enzyme 2 (ACE2, not shown) on the host’s cell surface. The fusion peptide (FP) fuses with the host cell membrane. Specific antibodies against RBD and FP can neutralize SARS-CoV-2 NTD/CTD, N-/C-terminal domains. (B) RBD is glycosylated and methylated, which may hinder the induction of neutralizing antibodies. In contrast, the receptor interaction site (RIS, green) is not glycosylated.
“Therefore, generating a vaccine inducing antibodies against RBD is the strategy used by the majority of COVID-19 vaccine candidates. It has recently been shown that RBD is glycosylated and methylated. Generally, such posttranslational modifications are difficult to reproduce in vaccines, meaning that vaccines may display (slightly) different epitopes than the virus. Consequently, the antibodies induced by the vaccines may potentially be cross-reactive and non-protective. Interestingly, however, the receptor interaction site (RIS) directly binding to ACE2 is not glycosylated, indicating that this RIS may potentially be an ideal vaccine candidate” [2] (Figures 4-7).
Figure 4: https://theconversation.com/COVID-19-vaccine-update-pfizer-may-be-the-frontrunner-but-canada-has-hedged-its-bets-149962
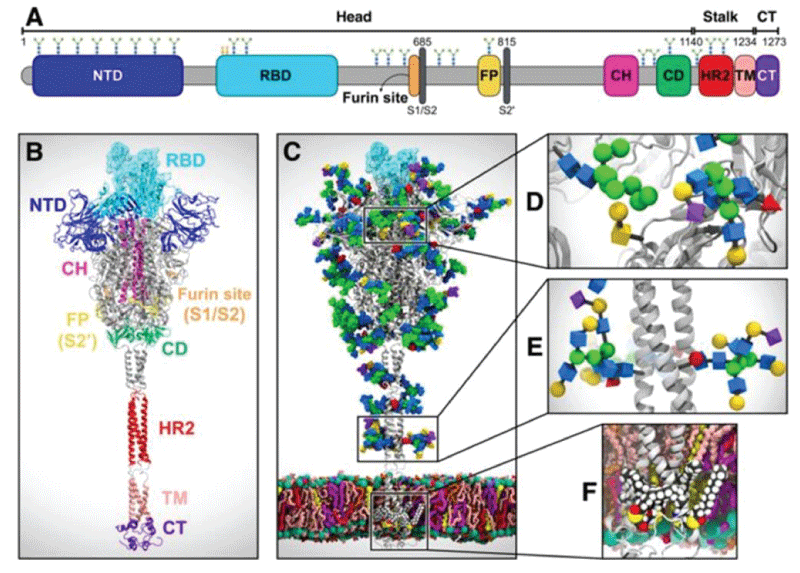
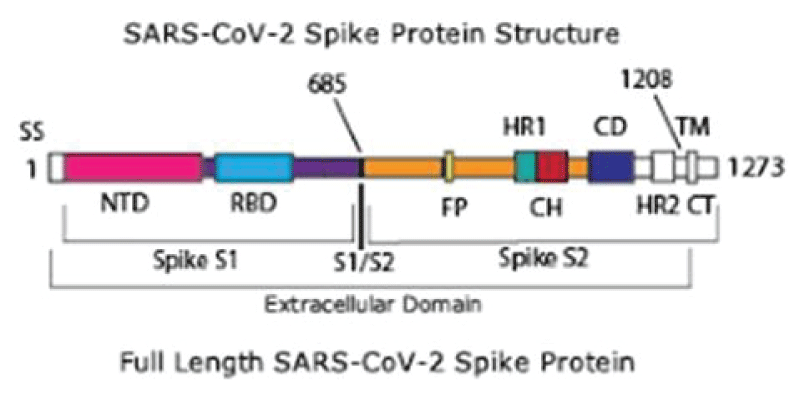
Figure 5: System overview. (A) A sequence of the full-length spike (S) protein contains the N-terminal domain (NTD, 16-291), receptor binding domain (RBD, 330-530), furin cleavage site (S1/S2), fusion peptide (FP, 788-806), central helix (CH, 987-1034), connecting domain (CD, 1080-1135), heptad repeat 2 (HR2, 1163-1210) domain, transmembrane domain (TD, 1214-1234), and cytoplasmic tail (CT, 1235-1273). Representative icons for N-glycans (blue and green) and O-glycan (yellow) are also depicted according to their position in the sequence. (B) Assembly of the head, stalk, and CT domains into a full-length model of the S protein. (C) Fully glycosylated and palmitoylated model of the Open system. (D-F) Magnified view of the N-/O-glycans rendered using the Symbol Nomenclature for Glycans (SNFG) (D, E) and S-palmitoylation of the cytoplasmic tail (F). from Casalino, et al.
Figure 6: According Immunogenicity of Candidate MERS-CoV DNA Vaccines Based on the Spike Protein.
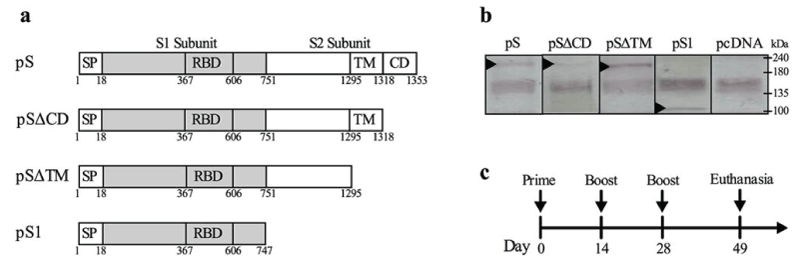
Figure 7: (a) Schematic representation of the generated DNA vaccine constructs. Four constructs were generated including one expressing full length S protein (pS) and three other constructs expressing truncated S protein with deleted cytoplasmic domain (pS∆CD), deleted transmembrane domain (pS∆TM) or deleted S2 subunit (pS1). Numbers indicate amino acids. SP: signal peptide; RBD: receptor-binding domain; TM: transmembrane domain; CD: cytoplasmic domain. (b) In vitro protein expression in cell culture. Vero E6 cells with 80% – 90% confluency were transfected with the DNA constructs; 48 h later, cell lysates were collected; protein expression was subsequently confirmed by western blot using anti-S1 polyclonal Abs. Arrows indicate band with expected molecular weight. (c) Time-line of immunization regimen.
Sawsan S. Al-amri, Ayman T. Abbas, Loai A. Siddiq, Abrar Alghamdi, Mohammad A. Sanki, Muhanna K. Al-Muhanna, Rowa Y. Alhabbab, Esam I. Azhar, Xuguang Li & Anwar M. Hashem.
Scientific Reports volume 7, Article number: 44875 (2017): “Several groups have investigated various vaccine platforms to combat MERS-CoV. Most of these experimental vaccines were based on MERS-CoV full-length or truncated versions of the spike protein; these prototype vaccines were found to have induced high levels of nAbs and sometimes conferred protection against MERS-CoV challenge in several animal models. However, several previous SARS-CoV vaccine studies have also shown that there might be some safety concerns associated with the use of WIV43, truncated S subunit/protein vaccines or vectored vaccines expressing full-length S protein45. These concerns included inflammatory and immunopathological effects such as eosinophilic infiltration of the lungs as well as Ab-mediated disease enhancement (ADE) in immunized animals upon viral challenge. It is believed that induction of Th2-polarized immune response and/or non-neutralizing Abs against epitopes within the S protein (i.e. outside the neutralizing-epitope rich RBD or S1 subunit) are the reason for the observed immunopathology and disease enhancement in vaccinated animals, suggesting that use of S1 subunit over full-length S protein could be a safer option for vaccine development.”
“The development of BNT162b1 and BNT162b2 started in Germany at BioNTech.
BNT162b1 encodes the trimerized (by addition of T4 fibritin folding domain) receptor binding-domain (RBD) of the spike-protein”.
“The development of BNT162b1 and BNT162b2 started in Germany at BioNTech.
BNT162b1 encodes the trimerized (by addition of T4 fibritin folding domain) receptor binding-domain (RBD) of the spike-protein” [3].
“vaccine astra Zeneca: The integrated gene encodes the full length spike-protein in its glycosylated form and includes a tissue plasminogen activator leader sequence “[3].
“our vaccine strategy based on the RBD has been supported by recent findings that most of the neutralizing activity is directed against the RBD in the sera of patients with COVID-19 and by evidence from human monoclonal antibodies [4].
More recently, Yang J, et al. have demonstrated that RBD-based COVID-19 vaccine could induce much higher titer of neutralizing antibody (NT50: ~2400) than S protein (NT50: ~300) and S1 subunit (NT50: ~1100).26 Currently, more than 150 COVID-19 vaccines are under development at different stages.6,7,8,9 Especially, a number of COVID-19 vaccines’ phase 1/2 clinical trials have been completed, including the adenovirus-vectored vaccines (Ad5-nCoV and ChAdOx1 nCoV-19) from CanSino10 and Oxford University/AstraZeneca,11 respectively; the mRNA vaccines (mRNA-1273 and BNT162b1) from Moderna12 and Pfizer/BioNTech,13 respectively; and the inactivated vaccines (PiCoVacc and BBIBP-CorV) from Sinovac14 and Beijing Institute of Biological Products,15 respectively (https://biorender.com/COVID-vaccine-tracker/). Generally speaking, all these vaccines could induce antibodies specific for spike (S) protein and receptor-binding-domain (RBD), which neutralized pseudotyped and live SARS-CoV-2 infection. Some reports have shown that the neutralizing antibody titers are strongly correlated with RBD-binding IgG concentration [5].
According a preprint: “Viral vector based, such as ChAdOx1 nCoV-19, as well as nucleic acid-based vaccine strategies, such as the Pfizer BNT162b2 and Moderna mRNA-1273 vaccines, rely on the supplied antigen-encoding DNA or RNA sequence, once inside a cell, to faithfully produce the spike-protein in its fully folded, glycosylated and assembled state, resembling a natural infection and trigger a robust innate -immune response, as well as provoking T and B cells. However, the cellular secretion pathway followed by such vaccine delivered antigens may differ in fundamental ways from antigens in the context of viral infection, where factors other than a single protein coding sequence may play decisive roles in immunogen presentation.
The Pfizer BNT162b2 vaccine antigen aims to overcome some of these important differences by following a strategy first employed for MERS, as well as SARS-CoV spike glycoprotein stabilisation for vaccine design, where two proline mutations are introduced in close proximity to the first heptad repeat of each protomer, which stabilises the prefusion conformation” [8].
“While this observation that the majority of cells infected with ChAdOx1 nCoV-19 present native-like spikes on the cell surface, it is interesting to note that a population may shed the S1 subunit. Whether this is a beneficial or detrimental feature with respect to the elicitation of immune responses during vaccination is unknown. Shedding of S1 subunits from viruses occurs during native infection, and the ChAdOx1 nCoV-19-derived S proteins mimic this native feature of the viral spike” [9].
According a preprint “The six COVID-19 vaccines currently in use around the world employ different strategies, and do not all incorporate the two proline substitutions that “lock” S into the pre-fusion conformer. Vaccines that do not utilize pre-fusion “locked” S are expected to produce lower levels of neutralizing antibodies, and hence may be less efficacious against infection, even if they do protect against severe COVID-19. Indeed, a two-dose regimen of the AstraZeneca ChAdOx1 based vaccine, which does not use a “locked” S, did not protect against mild-to-moderate COVID-19 in S. Africa, where 93% of COVID-19 cases in trial participants were caused by the B.1.351 variant. Like the AstraZeneca ChAdOx1 vaccine, the Sputnik V vaccine (Gam-COVID-Vac) is based on adenovirus vectored expression of a native S sequence, rather than a pre-fusion “locked” S. Although the Sputnik V vaccine has a reported vaccine efficacy of 91.6% in the interim analysis of Phase 3 trials held in Russia between Sept 7 and Nov 24, 2020, none of the VOC mentioned above nor independent lineages containing the E484K mutation were prevalent in Russia during this time period.
The heterogenous dose-response curves described is a property of Sputnik V vaccine elicited responses as soluble RBD-Fc inhibition of WT and VOC S-mediated entry produced classical dose response curves with Hill slopes close to -1.0.
RBD-Fc competition is an indirect measure of the virus spike affinity for human ACE2 expressed on the target F8-2 cell line. Both B.1.1.7 and B.1.351 were modestly but significantly more resistant to RBD-Fc inhibition. This is not surprising as both harbor the N501Y mutation known to enhance affinity of RBD for ACE2. However, this 1.5 – 2 fold increase in RBD-Fc IC50 for B.1.1.7 and B.1.351, respectively, does not explain the neutralization-resistant versus sensitive phenotype of B.1.351 versus B.1.1.7 in our virus neutralization assays.
Three of the six COVID-19 vaccines currently in use worldwide, namely Moderna mRNA-1273, BioNTech BNT162b2, and Janssen Ad26.COV2.S, each express S harboring K986P and V987P substitutions (2P) within a loop abutting the central helix of the S2’ membrane fusion machinery. This modification locks the spike in a prefusion conformation and elicits higher titers of neutralizing antibodies. Of the three vaccines that do not make use of 2P Spike mutants, Gamaleya’s Sputnik V and Astra Zeneca’s AZD1222 are adenovirus-vectored vaccines encoding native S. The third is Corona Vac, a preparation of inactivated SARS-CoV-2 virions. Although all six vaccines are highly efficacious at preventing severe COVID-19 outcomes, they do not all uniformly prevent infection. in all cases thus far examined, these first generation vaccines are less effective against variants with certain non-synonymous substitutions in Spike, such as E484K.
The most concerning variants are those with multiple mutations in the receptor binding domain (RBD) that confer both enhanced affinity for the hACE2 receptor and escape from neutralizing antibody responses.
B.1.351 and P.1 have in common three RBD substitutions (K417N/T, E484K and N501Y) whereas all three WHO designated VOC contain the N501Y substitution.
Although B.1.1.7 shows enhanced transmissibility and more severe disease outcomes, it does not appear to be particularly more resistant to serum neutralizing responses elicited by vaccines or natural infection. The same is not true, for the B.1.351 variant.
In live virus plaque reduction neutralization assays, sera from AstraZeneca vaccine recipients in South Africa exhibited 4.1 to 32.5 fold reduction in neutralizing activity against B.1.351 ” [10].
“Vaccine reactogenicity refers to a subset of inflammatory reactions that occur soon after vaccination. While no safety concerns appear to be related to the administration of Pfizer – BioNTech, Moderna, Oxford/AstraZeneca, and Gamalaya Res. Inst vaccine administration, the frequency of inflammatory reactions is higher than that commonly observed with flu vaccines. Of the two candidate vaccines initially tested by Pfizer – BioNThec, the BNT162b1 coding for the Receptor-Binding-domain (RBD) of the Spike-protein was dismissed since it was more reactogenic than the BNT162b2 coding for the entire Spike-protein. In the AstraZeneca trials, the ChAdOx1 candidate vaccine induced a similar immune response across all age groups, while it was less reactogenic in older adults than in younger adults. Early markers for reactogenicity, including the humoral innate immunity molecule PTX3, may help early assessment of activation of innate immunity and reactogenicity. COVID-19 has been associated in a fraction of patients with the development of autoimmune reactions blocking the Interferon pathway or eliciting thrombosis. Although there have been no major problems in patients suffering from autoimmune- disorders, further studies are needed to assess the impact of COVID-19 and vaccines on autoimmunity including myasthenia gravis” [12].
“The use of RBD in vaccines is compromised by its limited immunogenicity owing to its small molecular size and possible mixed forms of multiple complexes (as monomers, dimers or trimers). Strategies to overcome these drawbacks include increasing antigen size (for example, by fusing the RBD with an Fc domain) or by RBD multimerization (for example, by displaying multiple copies of RBD on particles)” [14].
Related the recent and interesting literature reported there are various kind of vaccine that use SPIKE-PROTEIN
Some use full length Spike-protein
Other use full length Spike-protein but with 2 mutation
Other are directed to RBD domain
And: “test for the presence of glycans, the team used HEK293 cells infected with the vaccine and used enzymes to generate glycopeptides. Tests revealed a high level of glycans present on the spike, providing evidence that the spike-proteins produced by ChAdOx1 vaccination were similar to the spike-protein expressed by natural infection, triggering an immune response that can protect”.
So some vaccine use the spike-proteins more similar to the spike-protein expressed by natural infection, triggering an immune response that can protect against COVID-19.
Because actually some regulative agency have reported the need to deeply investigate some of this new vaccine it is interesting to verify if the different target used can influence in some rare procoaugulant effect.
What can be the role played by 2 mutation on spike-protein or the role played by glicosilation?
A deeply molecular biology investigation is needed.
Because rare condition this effect is recognized with difficulty but it not must to be forgetted.
It is possible to consider a CLASS effect?
Future research will find the truth.
Ethical consideration: all international rules respected.
- Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2020; 21: 73–82. PubMed: https://pubmed.ncbi.nlm.nih.gov/33340022/
- Speiser DE, Bachmann MF. Vaccines COVID-19: Mechanisms of Vaccination and Immunity. Vaccines. 2020; 8: 404. PubMed: https://pubmed.ncbi.nlm.nih.gov/32707833/
- Prüβ BM. Review Current State of the First COVID-19 Vaccines. Vaccines 9010030. 2021; 9: 30. PubMed: https://pubmed.ncbi.nlm.nih.gov/33429880/
- Yang J, Wang W, Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020; 586: 572–577. PubMed: https://pubmed.ncbi.nlm.nih.gov/32726802/
- Liu Z, Xu W, Xia S, Gu C, Wang X, et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target Ther. 2020; 5: 282. PubMed: https://pubmed.ncbi.nlm.nih.gov/33247109/
- Zhang S, Liu Y, Wang X, Yang L, Li H, et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J Hematol Oncol. 2020; 13: 120. PubMed: https://pubmed.ncbi.nlm.nih.gov/32887634/
- Makowski L, Olson-Sidford W, Weisel JW. Biological and Clinical Consequences of Integrin Binding via a Rogue RGD Motif in the SARS CoV-2 Spike-protein. Viruses. 2021; 13: 146. PubMed: https://pubmed.ncbi.nlm.nih.gov/33498225/
- Brun J, Vasiljevic S, Gangadharan B, Hensen M, Chandran AV, et al. Analysis of SARS-CoV-2 spike glycosylation reveals shedding of a vaccine. 2020.
- Watanabe Y, Mendonça L, Allen RE, Howe A, Lee M, et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 nCoV-19/AZD1222. Vaccine. 2021. PubMed: https://pubmed.ncbi.nlm.nih.gov/33501433/
- Ikegame S, Siddiquey MNA, Hung CT, Haas G, Brambilla L, et al. Qualitatively distinct modes of Sputnik V vaccine-neutralization escape by SARS-CoV-2 Spike variants. medRxiv. 2021. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8020991/
- Folegatti PM. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomized controlled trial. 2020; 396: 467-478.
- Forni G, Mantovani A, COVID-19 Commission of Accademia Nazionale dei Lincei, Rome. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021; 28: 626–639. PubMed: https://pubmed.ncbi.nlm.nih.gov/33479399/
- Luisetto M, Almukhtar N, Hamid GA, Khan FA. Spike sars-cov-2 protein as procoagulant factor and vaccine class effect hypothesis. 2021. https://www.researchgate.net/publication/350721725_SPIKE_SARS-COV-2_PROTEIN_AS_PROCOAGULANT_FACTOR_AND_VACCINE_CLASS_EFFECT_HYPOTHESIS
- Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2020; 21: 73–82. PubMed: https://pubmed.ncbi.nlm.nih.gov/33340022/
- Tobaiqy M, Elkout H, MacLure K. Analysis of Thrombotic Adverse Reactions of COVID-19 AstraZeneca Vaccine Reported to Eudra Vigilance Database. Vaccines. 2021; 9: 393.
- Al-amri SS, Abbas AT, Siddiq LA, Alghamdi A, Sanki MA, et al. Immunogenicity of Candidate MERS-CoV DNA Vaccines Based on the Spike Protein. Scientific Reports. 2021; 7: 44875.